Welcome to SpikeInterface’s documentation!
SpikeInterface is a Python module to analyze extracellular electrophysiology data.
With a few lines of code, SpikeInterface enables you to load and pre-process the recording, run several state-of-the-art spike sorters, post-process and curate the output, compute quality metrics, and visualize the results.
Overview of SpikeInterface modules
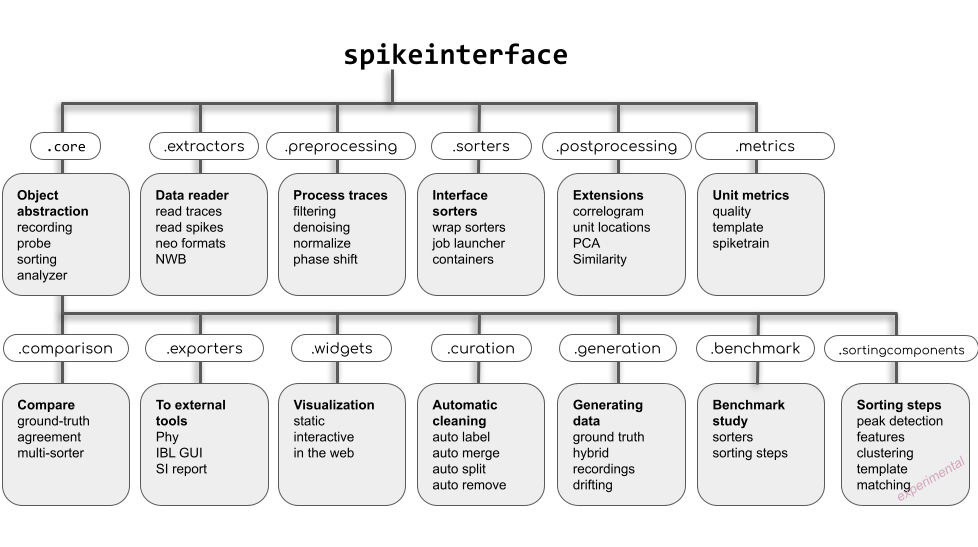
SpikeInterface is made of several modules to deal with different aspects of the analysis pipeline:
read/write many extracellular file formats.
pre-process extracellular recordings.
run many popular, semi-automatic spike sorters (kilosort1-4, mountainsort4-5, spykingcircus, tridesclous, ironclust, herdingspikes, yass, waveclus)
run sorters developed in house (lupin, spkykingcicus2, tridesclous2, simple) that compete with kilosort4
run theses polar sorters without installation using containers (Docker/Singularity).
post-process sorted datasets using th SortingAnalyzer
compare and benchmark spike sorting outputs.
compute quality metrics to validate and curate spike sorting outputs.
visualize recordings and spike sorting outputs in several ways (matplotlib, sortingview, jupyter, ephyviewer)
export a report and/or export to phy
curate your sorting with several strategies (ml-based, metrics based, manual, …)
offer a powerful Qt-based or we-based viewer in a separate package spikeinterface-gui for manual curation that replace phy.
have powerful sorting components to build your own sorter.
have a full motion/drift correction framework (See Motion/drift correction)
Other resources
To get started with SpikeInterface, you can take a look at the following additional resources:
- spiketutorials is a collection of basic and advancedtutorials. It includes links to videos to dive into the SpikeInterface framework.
- SpikeInterface Reports contains several notebooks to reproduce analysisfigures of SpikeInterface-based papers and to showcase the latest features of SpikeInterface.
- The 2020 eLife paper introduces the concept and motivation andperforms an in-depth comparison of multiple sorters (spoiler alert: they strongly disagree with each other!).Note: the code-base and implementation have changed a lot since the “paper” version published in 2020.For detailed documentation we therefore suggest more recent resources, like this documentation and
spiketutorials.